Continuous Matrix Factorizations
and their relation to kernel approximations
Introduction
This note was inspired by the plenary talk by Joel A. Tropp at SIAM LA24. While I could not find the specific talk online a very similar talk was recorded at the Boeing Colloquium Series. In the talk Joel presented the work of
The (Discrete) Cholesky Factorization
The Cholesky factorization of a positive semidefinite matrix $\mathbf{K}$ is given by
\[\mathbf{K} = \mathbf{L}\mathbf{L}^\top \in \mathbb{R}^{n\times n},\]where $\mathbf{L}$ is a lower triangular matrix. Where $\mathbf{L}$ and $\mathbf{L}^\top$ are the factors of interest. The factorization is useful as it can be used to solve the linear systems $\mathbf{K}\mathbf{s} =\mathbf{t}$ by performing two triangular solves ($\mathbf{L}\mathbf{y} = \mathbf{t}, \mathbf{L}^\top\mathbf{s} = \mathbf{y}$). The Cholesky factorization can be computed by performing $n$ rank-1-updates of the original matrix. The first iterate is
\[\mathbf{K}^{(0)} = \begin{bmatrix}k & \mathbf{m}^\top \\ \mathbf{m} & \mathbf{M}\end{bmatrix} = \frac{1}{k}\begin{bmatrix}k \\ \mathbf{m}\end{bmatrix}\begin{bmatrix}k \\ \mathbf{m}\end{bmatrix}^\top + \underbrace{\begin{bmatrix} 0 & 0 \\0 & \mathbf{M} - \mathbf{m}\mathbf{m}^\top /k\end{bmatrix}}_{\text{Residual. Denoted by } \mathbf{K}^{(1)}}.\]Each iteration eliminates a row and a column of the residual $\mathbf{K}^{(i)}$, with the first residual being the matrix itself. Thus after $r$ rank-1-update we have eliminated $r$ rows and columns of $\mathbf{K}$. The rows and columns that gets eliminated at the $i$th iteration is called the $i$th pivot. After $n$ iterations every row and column have been eliminated and we have the full factorization.
In this note we will mostly be interested in the partial Cholesky factorization, which corresponds to stopping the Cholesky factorization after $r$ iterations. The partial Cholesky factorization will be a rank $r$ approximation of $\mathbf{K}$. The aim is that if $r \ll n$, then the approximation is a “data efficient” representation of $\mathbf{K}$. There exist various of approaches of how to chose the pivots
Pivoting strategies
Let $\mathcal{I}_i$ be the unpicked columns after $i$ iterations. Then we can pick the next pivots as
-
Greedy: Pick the Next pivot element on the diagonal \(s_i = \text{argmax}_{k\in \mathcal{I}_i}\ \mathbf{A}^{(i-1)}(k,k)\)
-
Uniform: Pick uniformly \(s_i \sim \text{uniform}\{\mathcal{I}_i\}\)
-
Random pivoting: Pick with probability proportional to the diagonal element \(\mathbb{P}\left\{s_i = j\right\} = \frac{\mathbf{A}^{(i-1)}(j,j)}{\text{tr}\ \mathbf{A}^{(i-1)}}, \quad \forall j = 1,\dots,N\)
Note that the above strategies are all the same family of choosing the pivots w.r.t. the Gibbs distribution (the above can be achieved by $\beta \in \lbrace\infty,0,1\rbrace$)
\[\mathbb{P}\left\{s_i = j\right\} = \frac{|\mathbf{A}^{(i-1)}(j,j)|^\beta}{\sum_{k=1}^N |\mathbf{A}^{(i-1)}(k,k)|^\beta}, \quad \forall j = 1,\dots,N\]In
Kernel Matrices and Kernel Functions
The motivation for the note is that of Gaussian process regression. A key concept in Gaussian process regression is that of a kernel matrix. The kernel matrix, $\mathbf{K}$, is a matrix whose elements are generated from a so-called kernel function $k(\mathbf{x},\mathbf{y})$.
\[\mathbf{K}_{ij} = k(\mathbf{x}_i, \mathbf{x}_j),\ \text{ where }\ \mathbf{x}_i, \mathbf{x}_j \in X\]where $X = \lbrace\mathbf{x}_1, \mathbf{x}_2, \dots, \mathbf{x}_n \rbrace$ is a collection of data points. An important property of the kernel function is that it is positive semidefinite
\[k(\mathbf{x},\mathbf{x}) \geq 0,\]which has the consequence of the resulting kernel matrix $\mathbf{K}$ is positive semidefinite. A commonly used kernel function is the squared exponential given by
\[k(\mathbf{x},\mathbf{x}) = \exp\left(-\frac{\|\mathbf{x}_i - \mathbf{x}_j\|_2^2}{2\ell}\right),\]where $\ell$ is commonly referred to as the length scale.
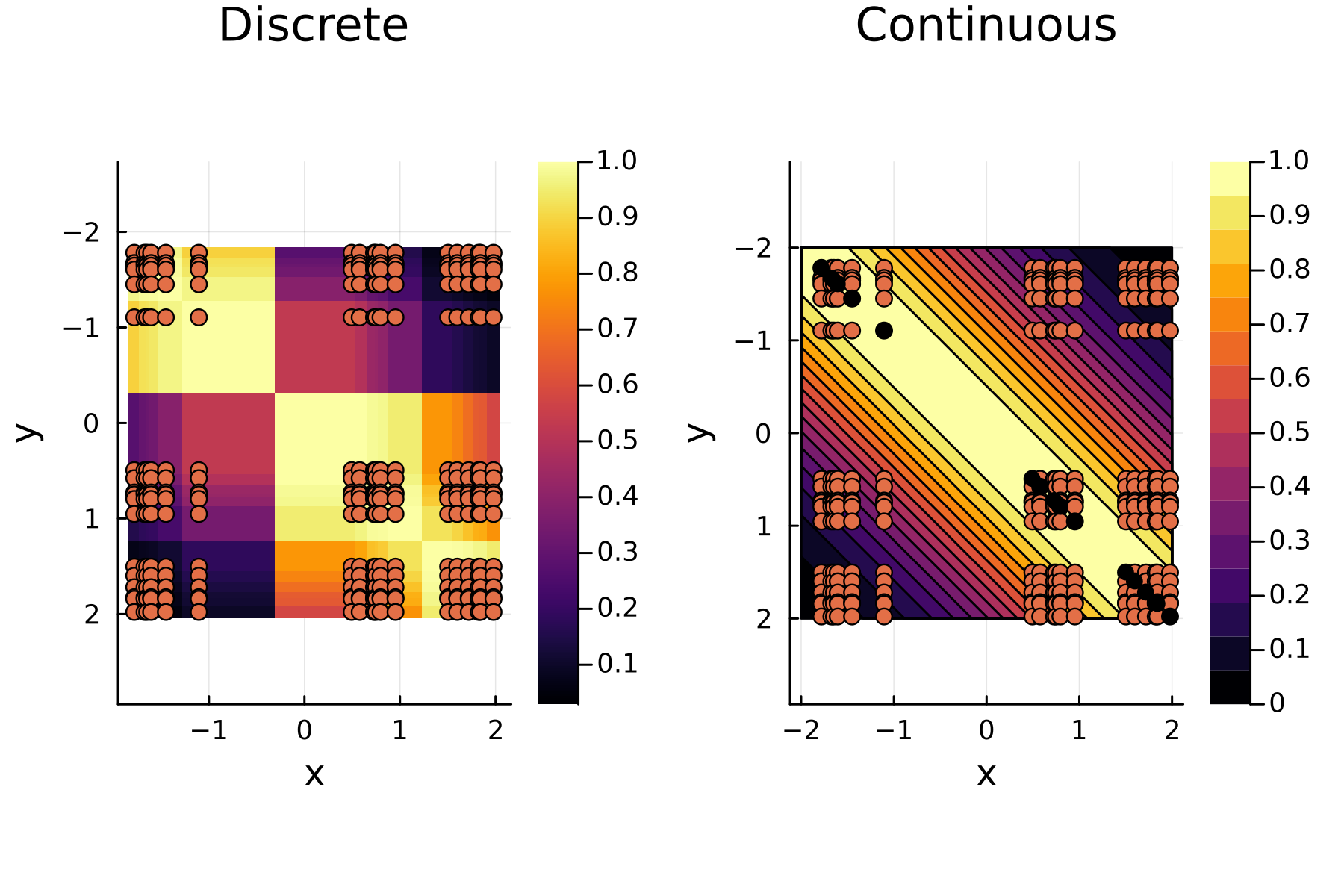
To illustrate the process we simulate data points $\mathbf{x}_i \in [-2,2]$ and plot both the discrete / observed kernel matrix $\mathbf{K}$ and the continuos kernel function. Two things to note: First that the $y-$axes below are flipped to conform to the standard kernel matrix form and secondly that the data is sorted, giving some structure to the kernel matrix.
using LinearAlgebra, StatsBase, Plots, Random, InvertedIndices
Random.seed!(1234)
n = 20 # Number of data points
k = 5 # Rank of approximation
dim = 1 # Dimension of data
l = 2.0 # Lengthscale parameter
X = [rand(floor(Int,n/3),dim) .- 2; rand(floor(Int,n/3),dim); rand(floor(Int,n/3),dim)/2 .+ 1.5] # Three groups
n = length(X) # Number of data points
X = sort(X[:]) # Soring the data - Not strictly necessary, but makes the discrete form nicer
G(x,y,l=l) = exp(-norm(x-y)^2/(2*l))
# The actual view is in 2D
o = ones(n)
Xx = kron(X,o)
Xy = kron(o,X)
Gk = [G(x,y) for (x,y) in zip(Xx,Xy)]
plot_matrix = heatmap(sort(X[:]),sort(X[:]),reshape(Gk,n,n),aspect_ratio=:equal, title="Discrete")
scatter!(plot_matrix,Xx,Xy, label=false)
xlabel!(plot_matrix,"x"); ylabel!(plot_matrix,"y"); yflip!(true)
Xc = range(-2,2,300)
plot_smooth = contour(Xc,Xc, (x,y) -> G(x,y), fill=true,aspect_ratio=:equal,clim=(0,1))
xlabel!(plot_smooth,"x"); ylabel!(plot_smooth,"y"); yflip!(true)
scatter!(plot_smooth,Xx,Xy, label=false,title="Continuous")
scatter!(plot_smooth,X,X, label=false, color=:black)
plot(plot_matrix,plot_smooth, layour=(1,2),dpi=300)
The Continuous Cholesky Factorization
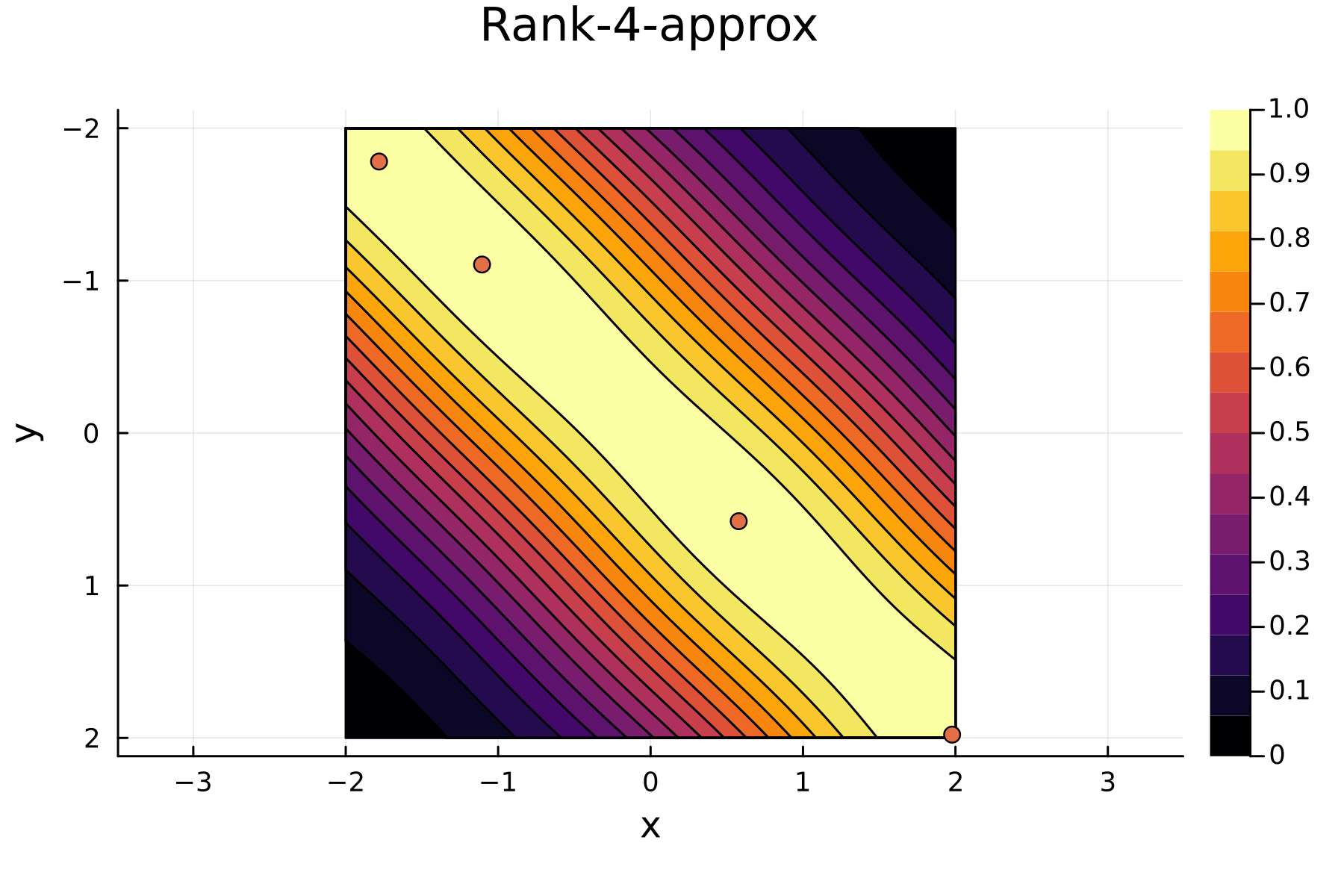
Approximating the discrete form with a low-rank matrix is as easy performing a partial Cholesky. However, a different approach is needed in order to approximate the continuous form of the “low-rank” function. In
The continuous Cholesky factorization of rank $r$ of kernel function is given as
\[k(\mathbf{x},\mathbf{y}) \approx \sum_{i=1}^{r}\frac{k_i(\mathbf{x},\mathbf{x}_i)k_i(\mathbf{x}_i, \mathbf{y})}{k_i(\mathbf{x}_i, \mathbf{x}_i)} = \sum_{i=1}^{r}\frac{k_i(\mathbf{x},\mathbf{x}_i)}{\sqrt{k_i(\mathbf{x}_i, \mathbf{x}_i)}}\frac{k_i(\mathbf{x}_i, \mathbf{y})}{\sqrt{k_i(\mathbf{x}_i, \mathbf{x}_i)}}\]where $(\mathbf{x}_i,\mathbf{x}_i)$ are the so-called pivot points and
\[k_i(\mathbf{x},\mathbf{y}) = \begin{cases} k(\mathbf{x},\mathbf{y}) \quad &i = 1\\ k_{i-1}(\mathbf{x},\mathbf{y}) - \frac{k_{i-1}(\mathbf{x},\mathbf{x}_{i-1})k_{i-1}(\mathbf{x}_{i-1},\mathbf{y})}{k_{i-1}(\mathbf{x}_{i-1},\mathbf{x}_{i-1})}\quad &i \geq 2. \end{cases}\]Visual Representations
The Greedy Approach
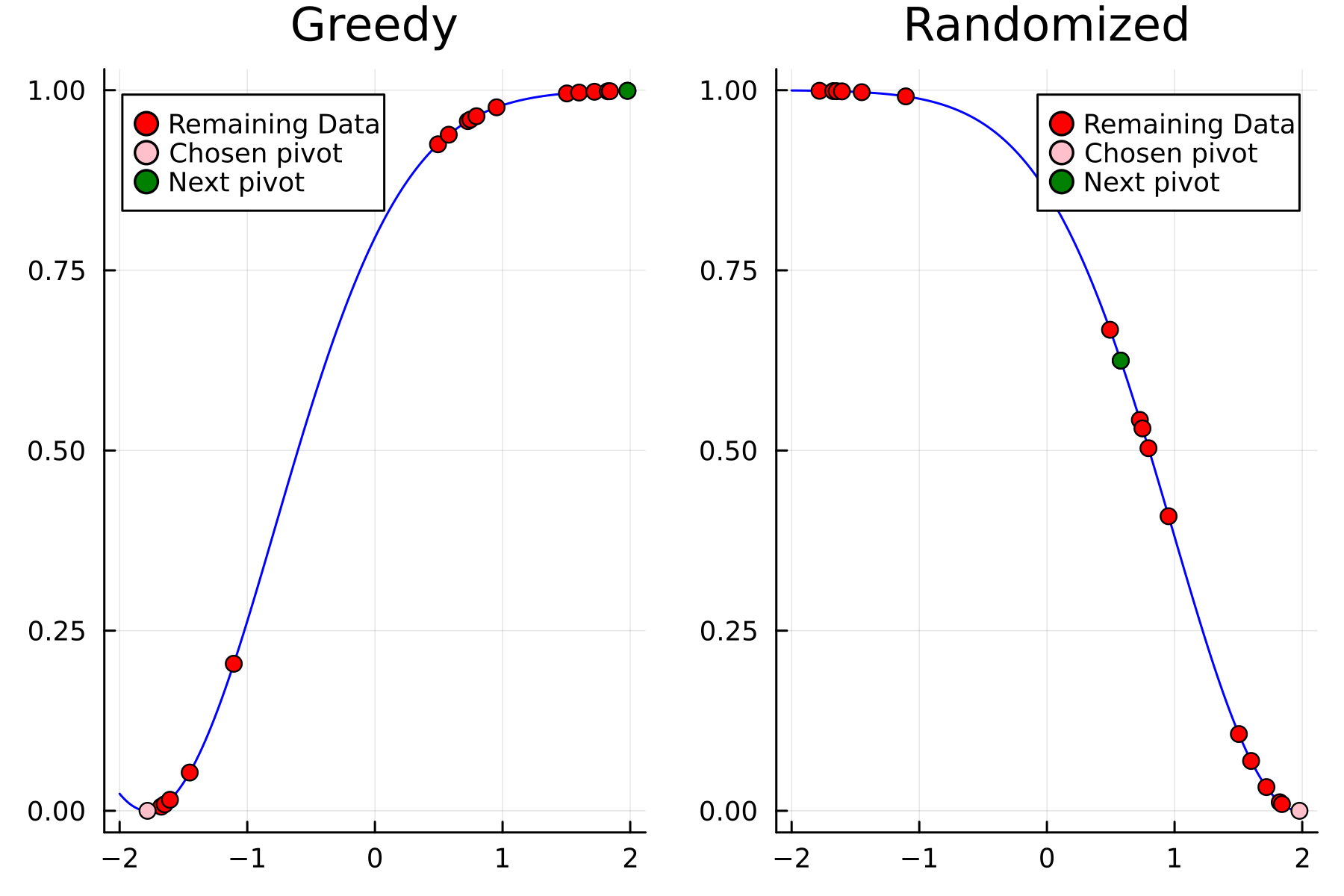
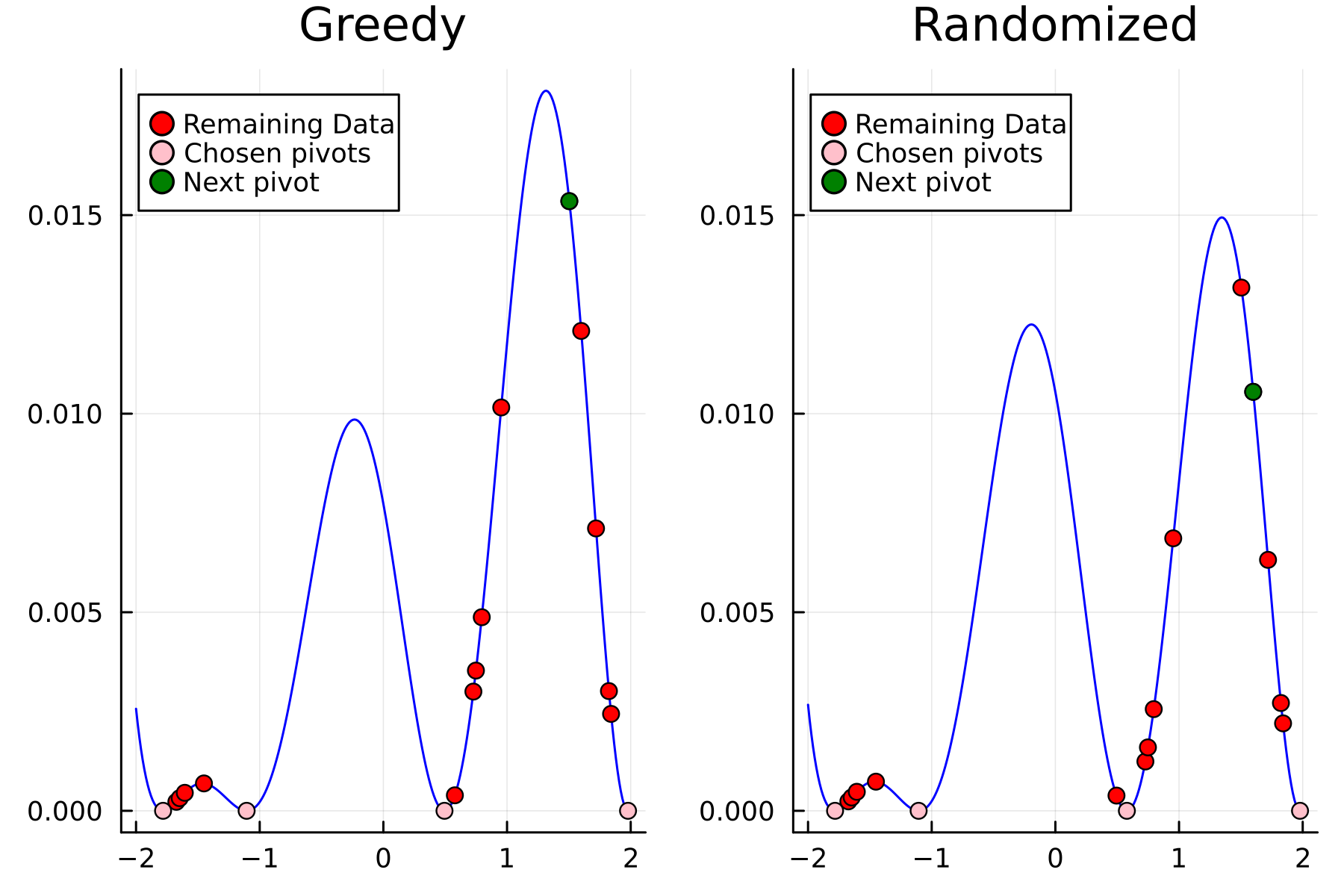
As the initial residual is just the matrix itself $(\mathbf{K}^{(0)})$ then its diagonal is filled with ones. As such the greedy method will simply pick the first pivot depending on the ordering of the data. This is one of the weaknesses of the greedy method, namely that is susceptible to bad ordering of the data. In fact
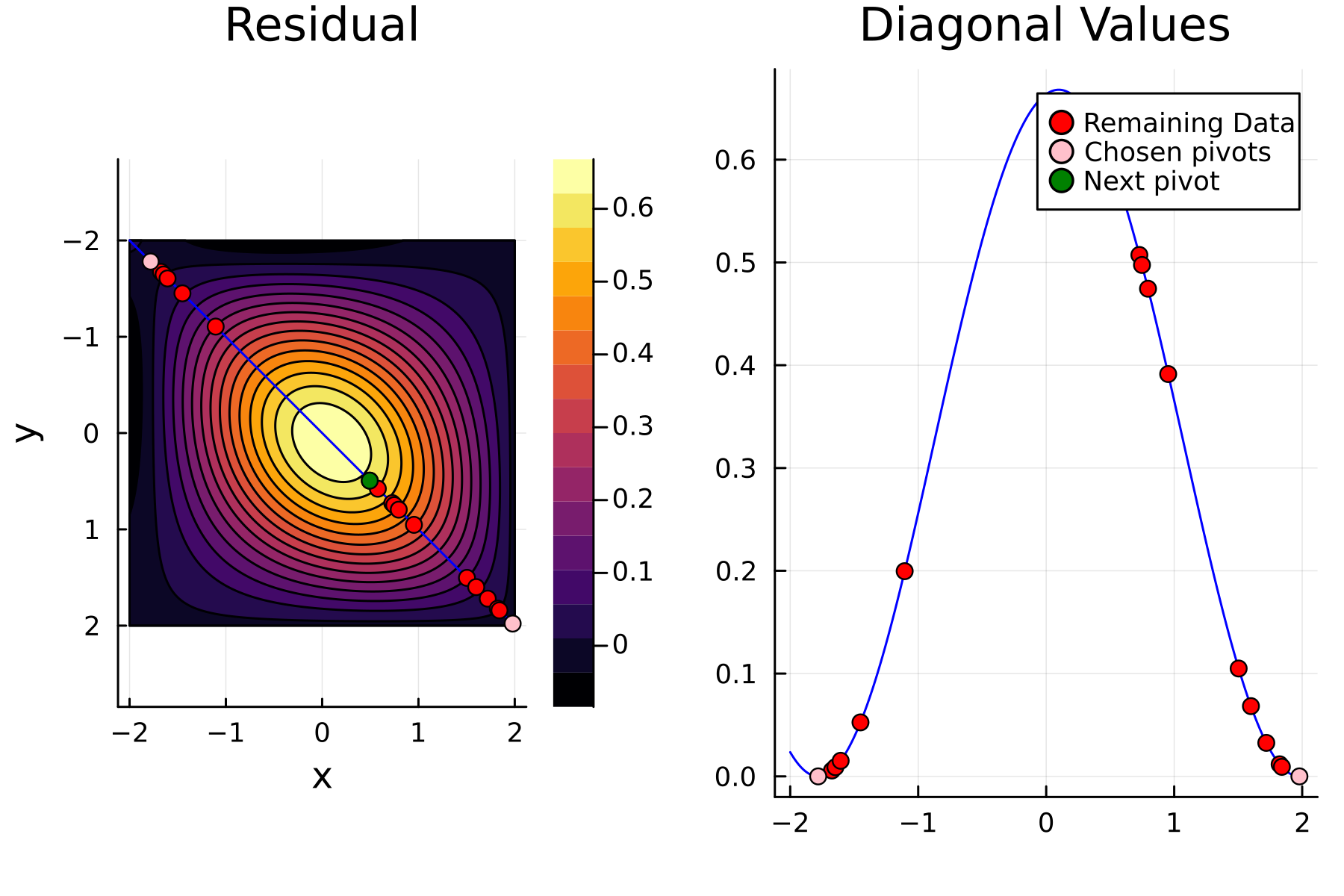
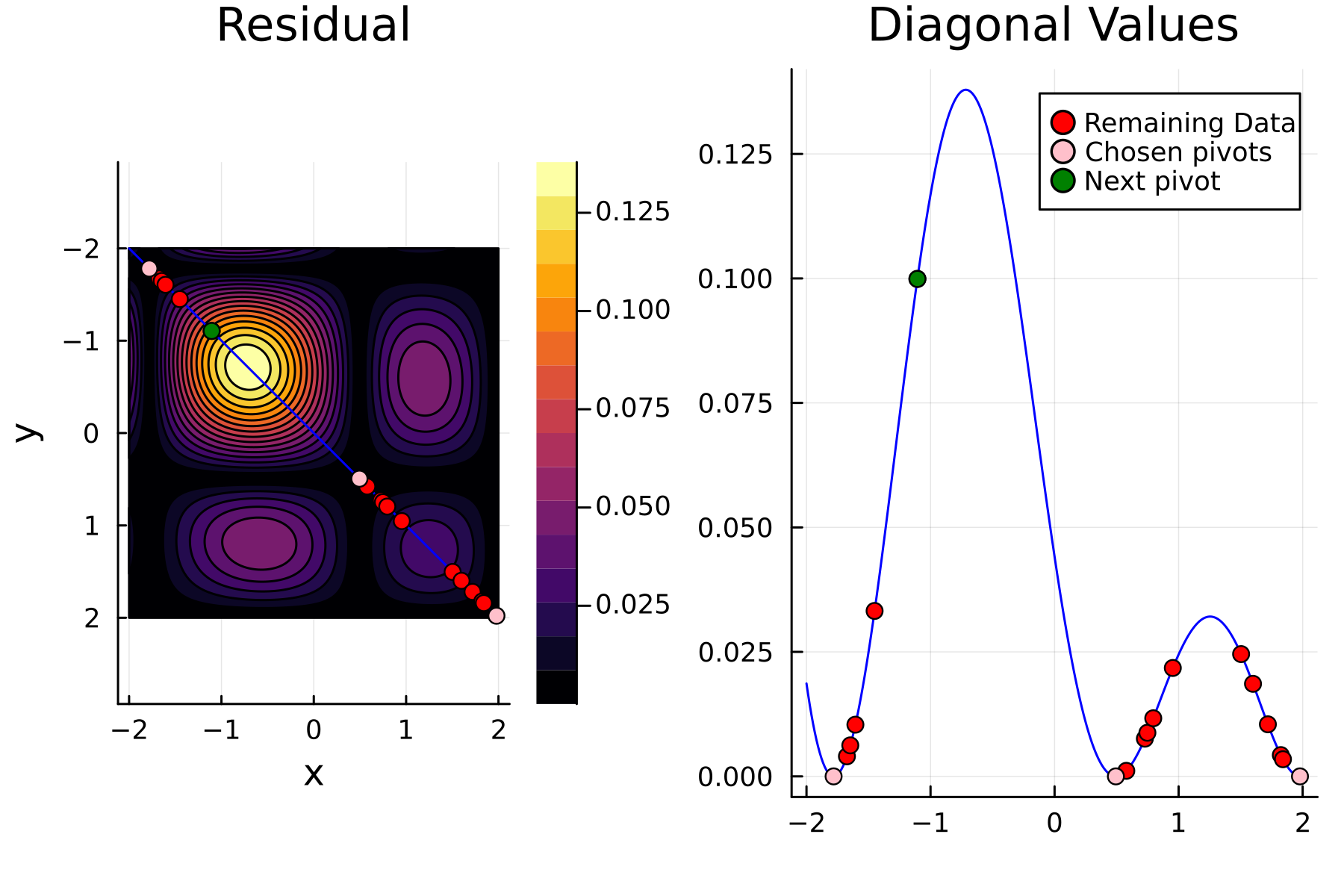
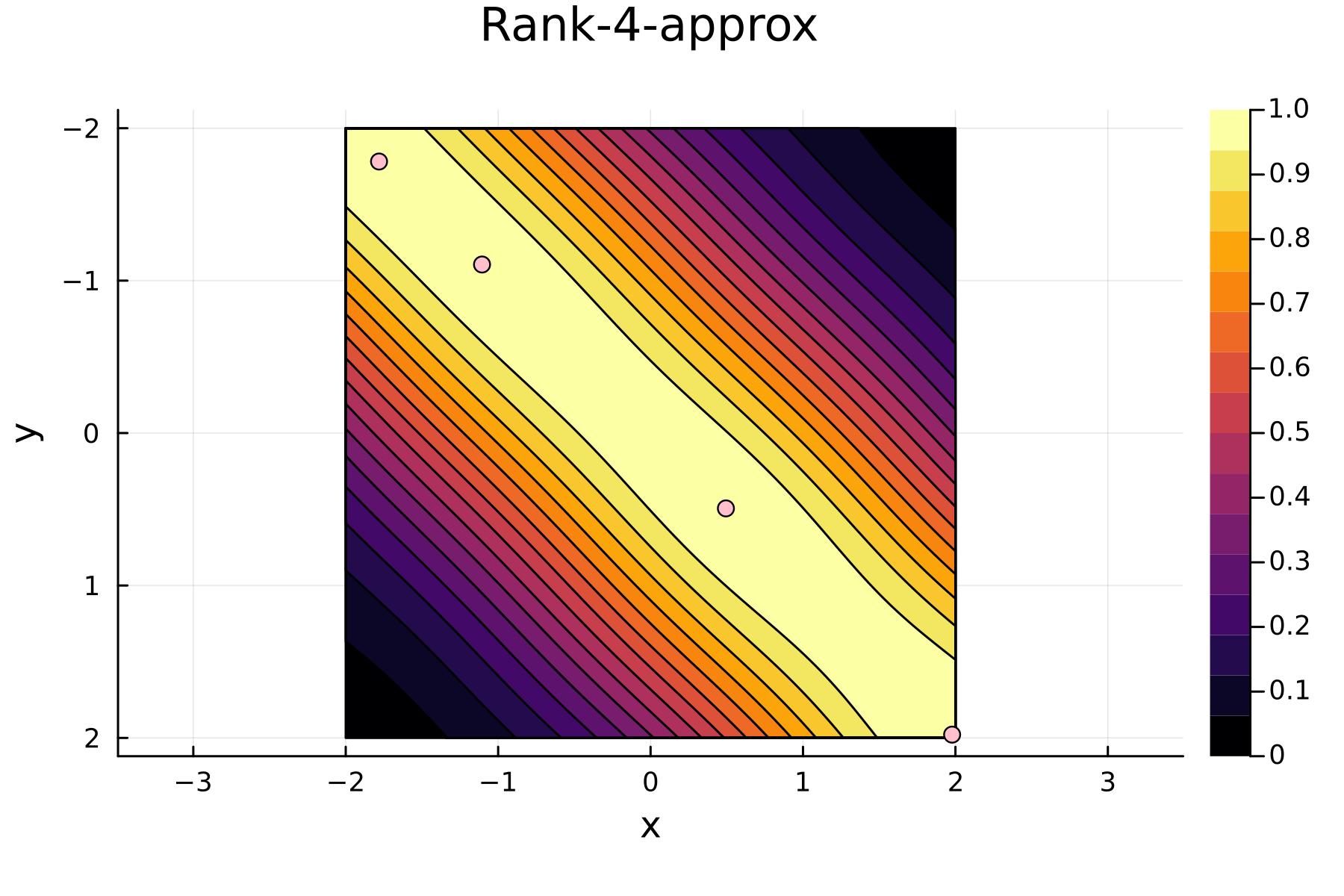
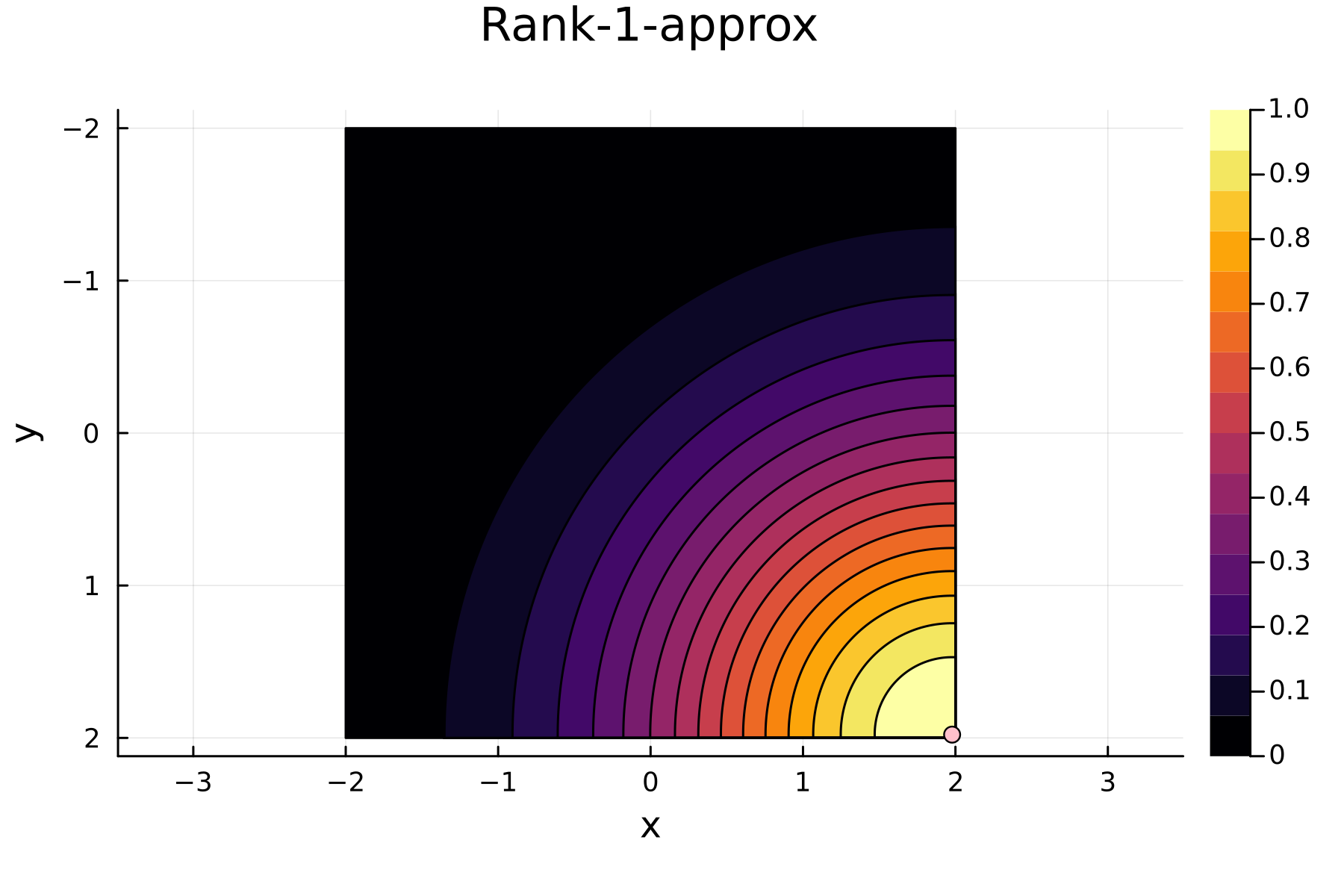
In the previously data introduces is completely ordered w.r.t. $x$, and the greedy method therefore picks the first pivot as the point with smallest value. The next iteration again start by looking at the diagonal, but this time of the residual $\mathbf{K}^{(1)}$. Given that the first pivot was chosen as smallest value, it is no surprise that the worst approximation happens furthest away. As such the next pivot point will be at the largest value. The two iterations highlight the weakness of the greedy method: In most cases it ends up picking pivots on the borders of the dataset.
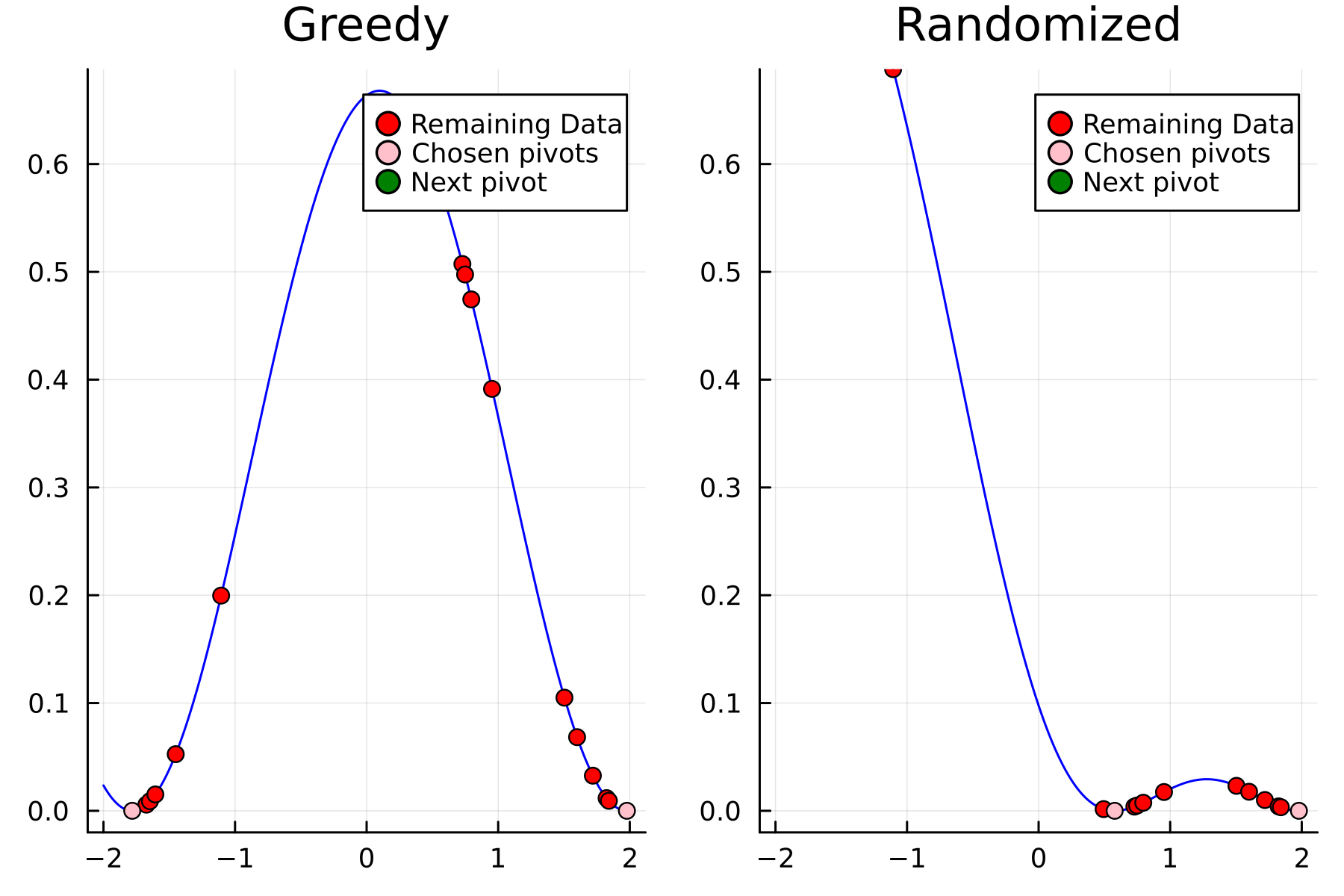
The series of the first 4 iterations can be seen below.
1st iteration
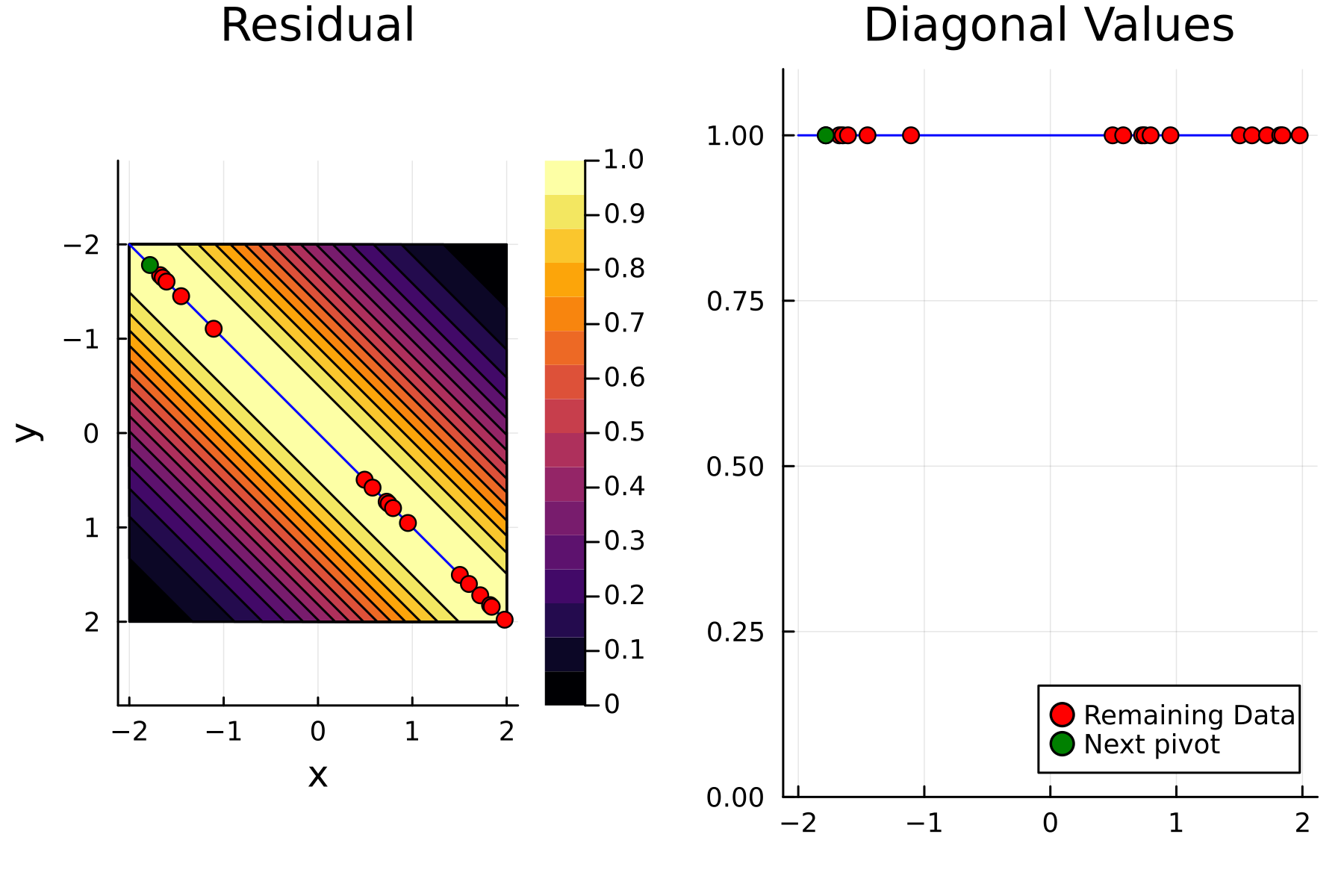
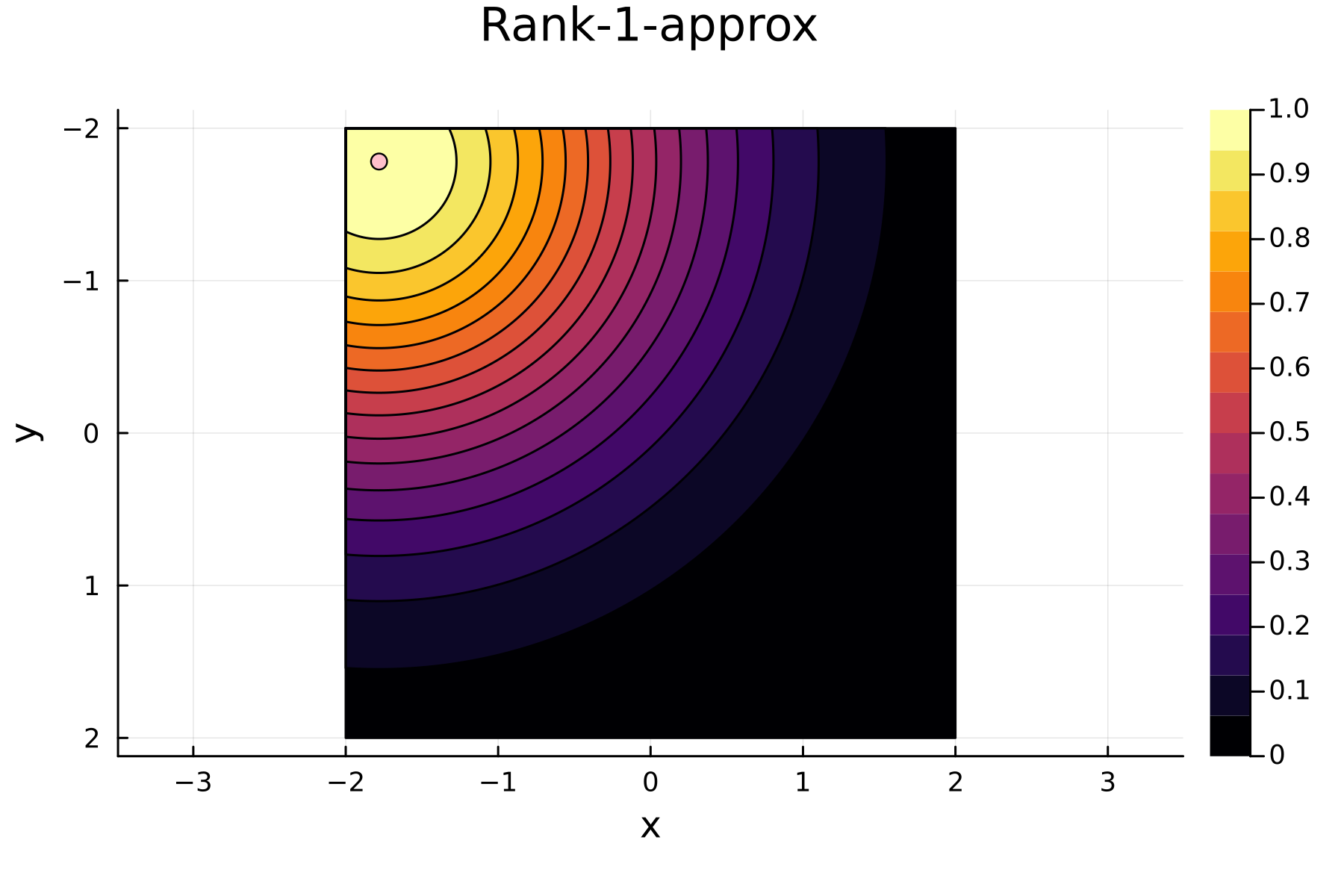
2nd iteration
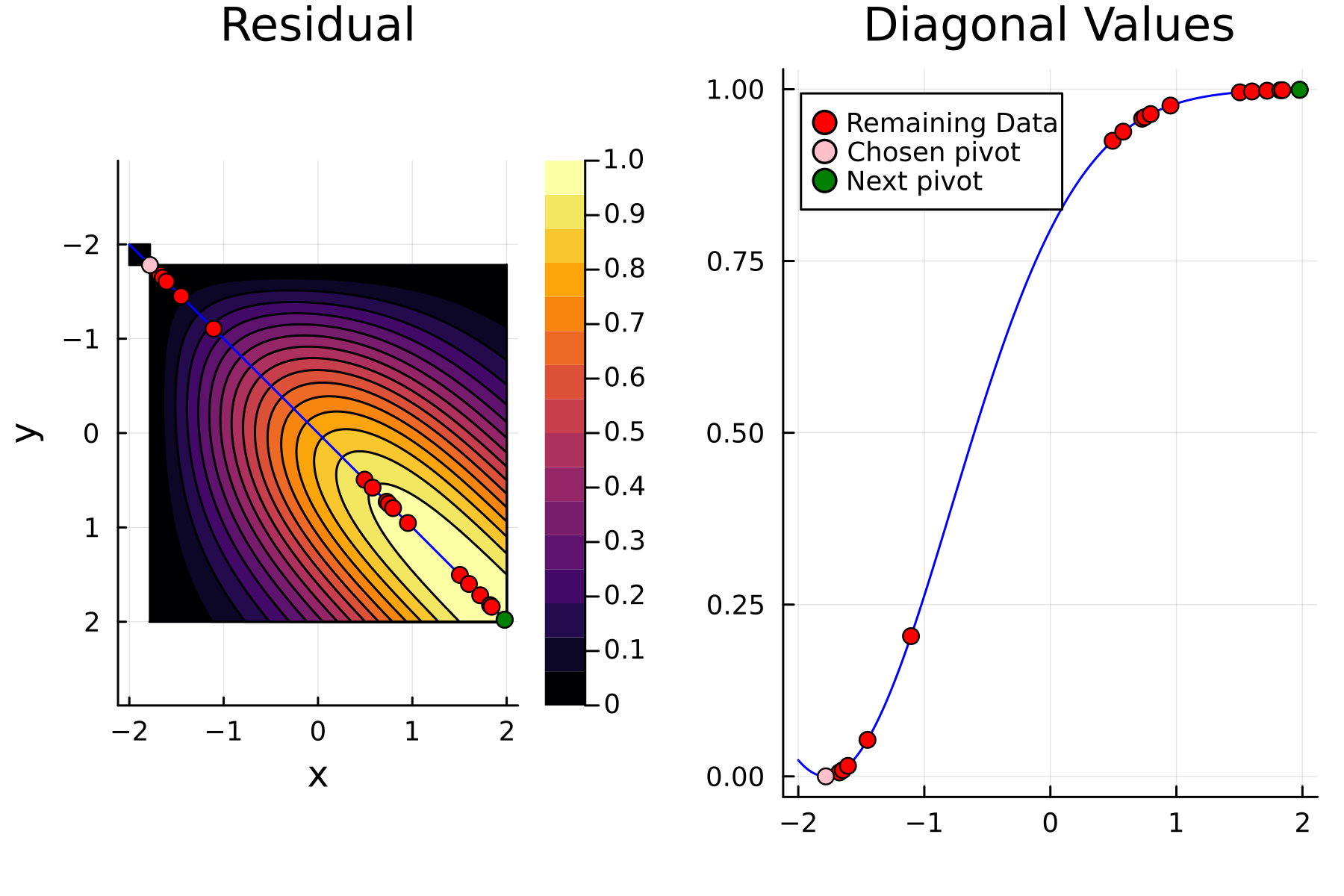
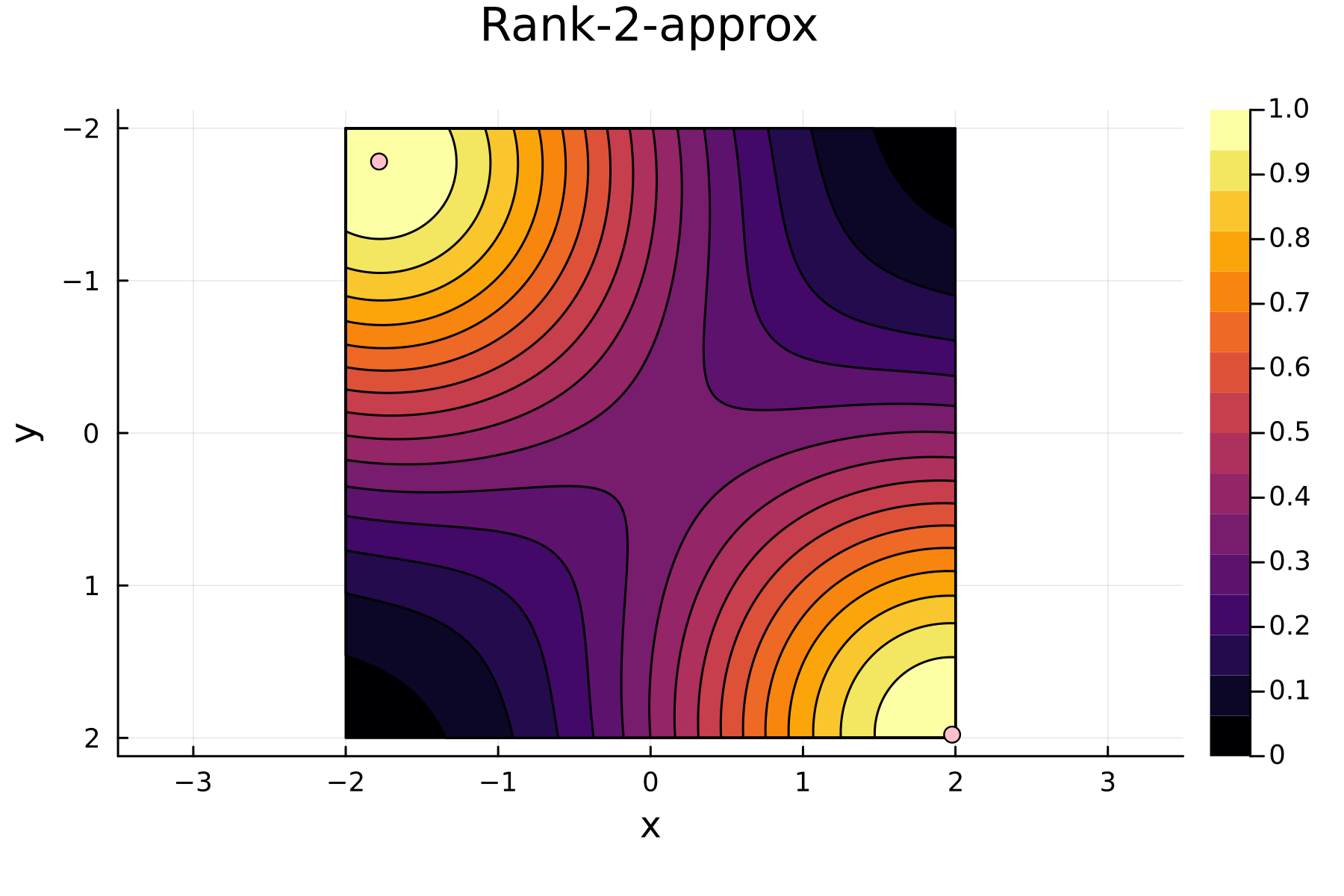
3rd iteration
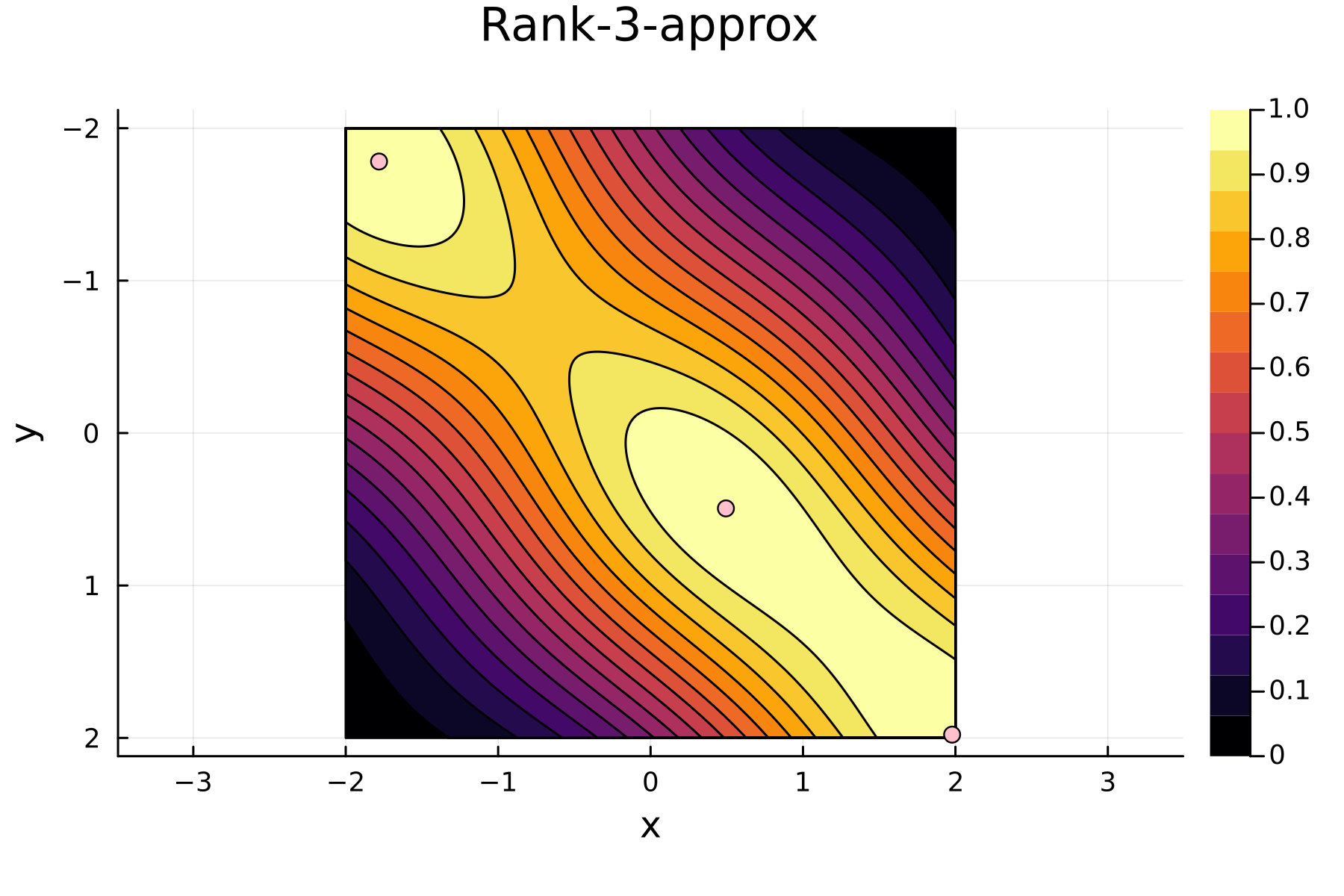
4th iteration
The Randomized Approach
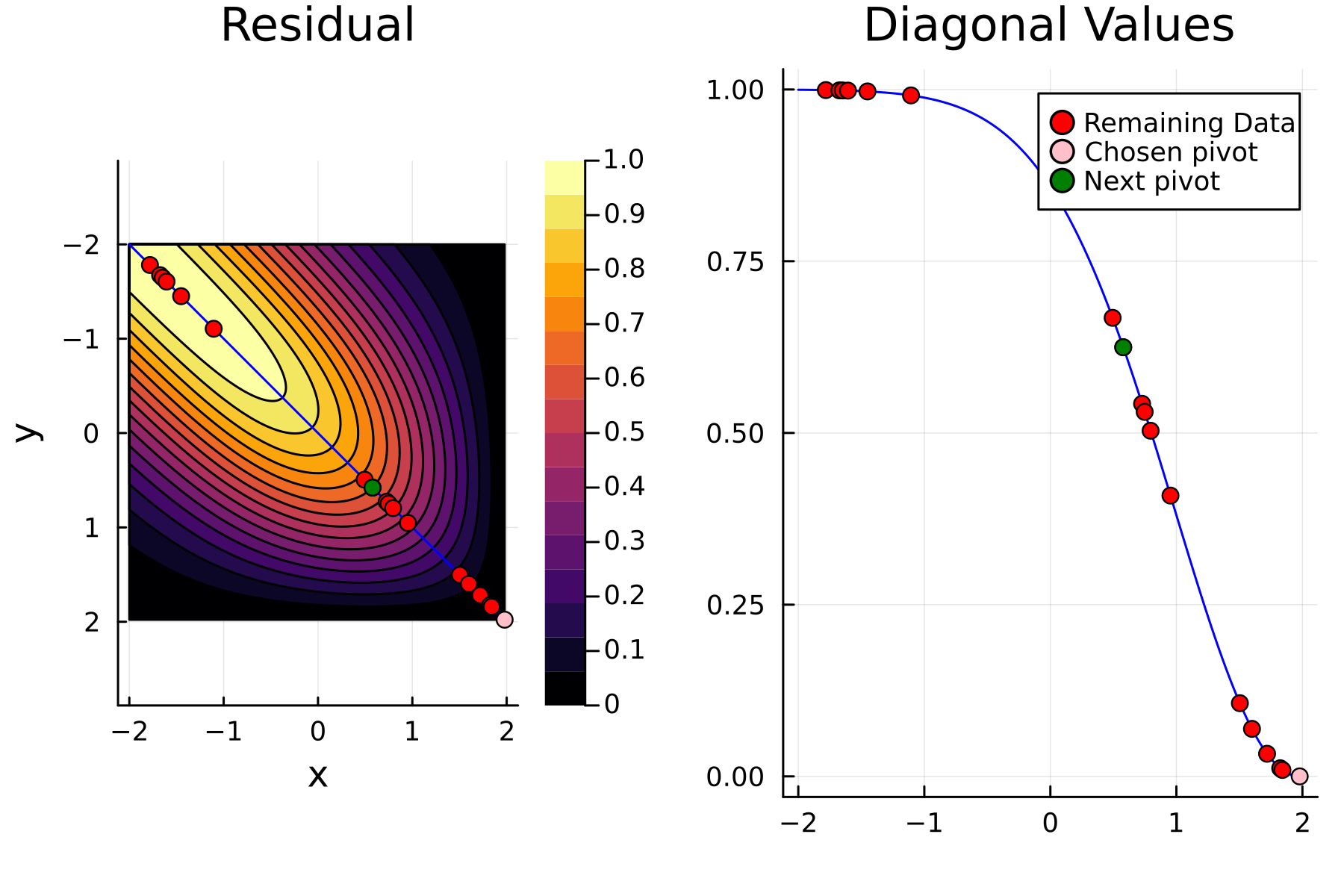
We now repeat the first 4 steps, but using randomized pivoting approach. A noticeable difference between the greedy and the randomized approach is that the randomized approach prioritizes (with some probability off course) to pick points close to where the data actually is. This is a win, as a point “inside” of the dataset will reduce the overall diagonal of the residual in the places of where data is actually located. In addition, the approach is also completely robust against the ordering of the data, as a randomization of the data is built directly into the approach.
1st iteration
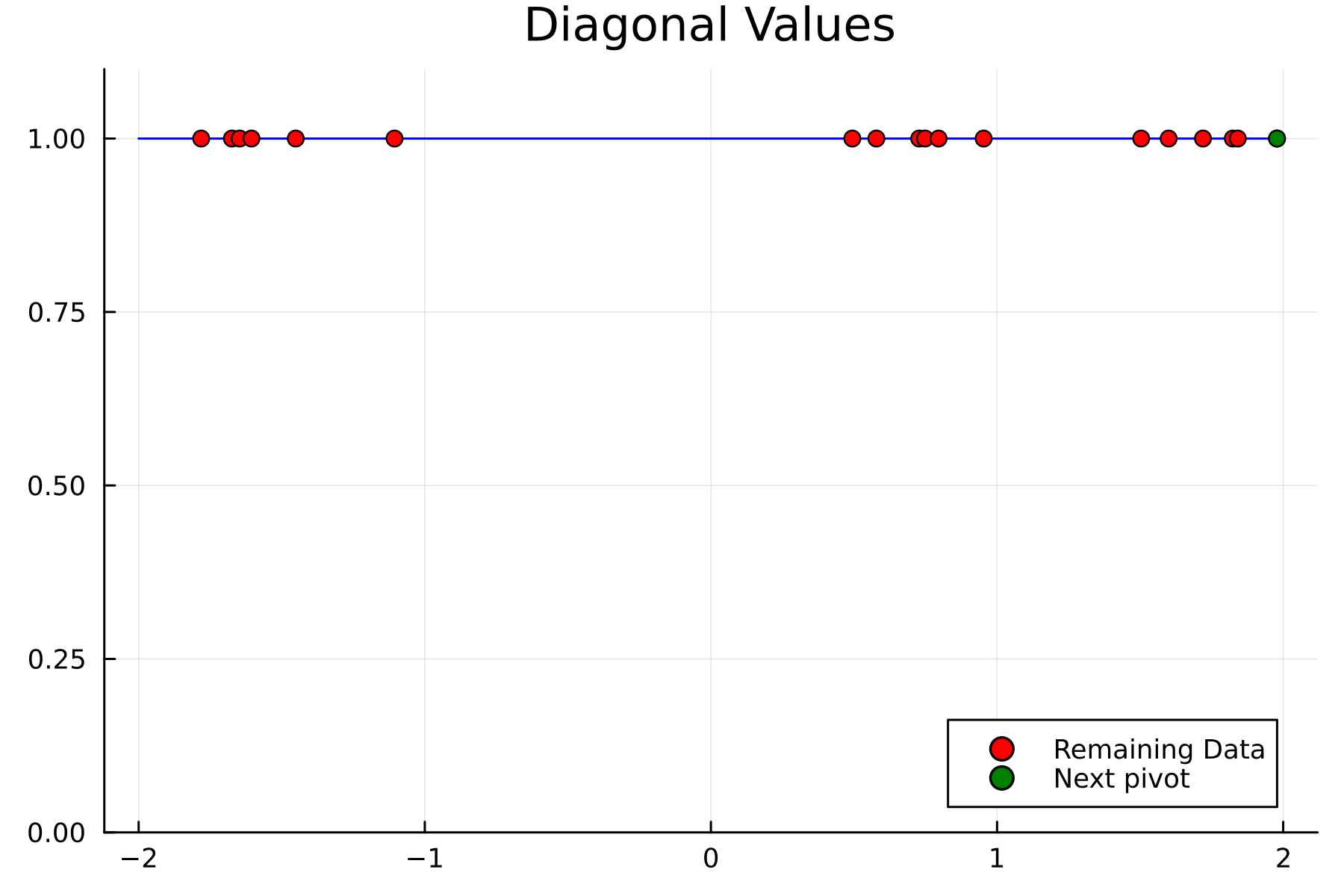
2nd iteration
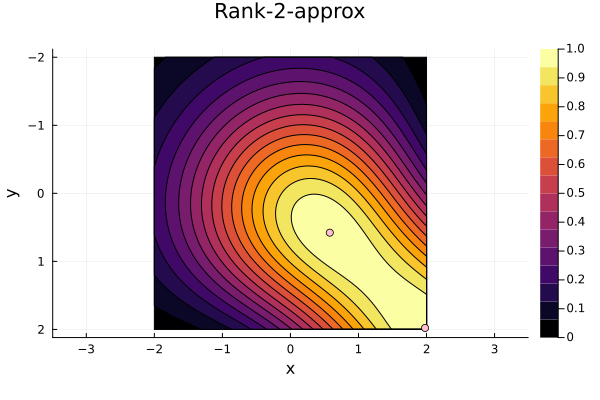
3rd iteration
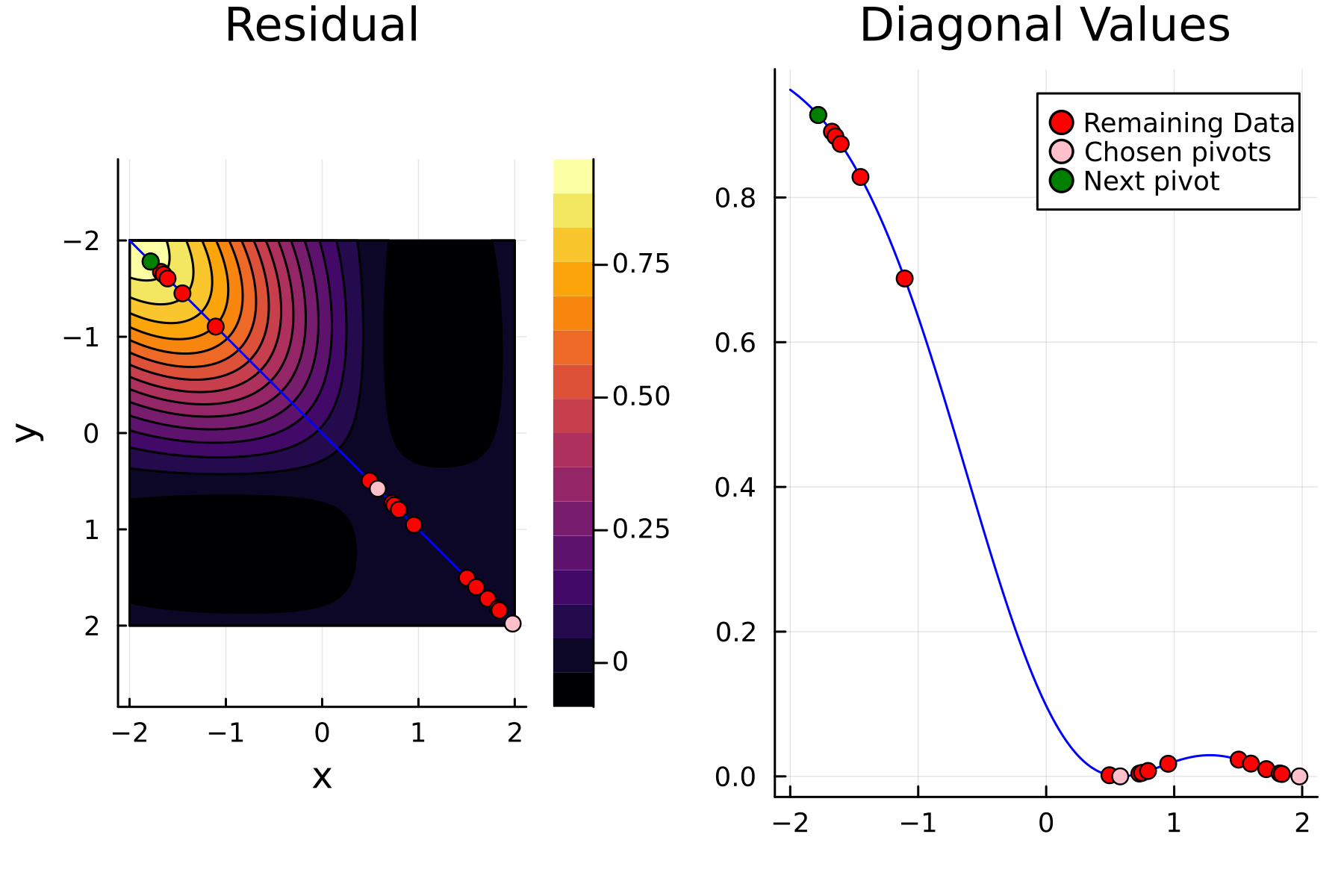
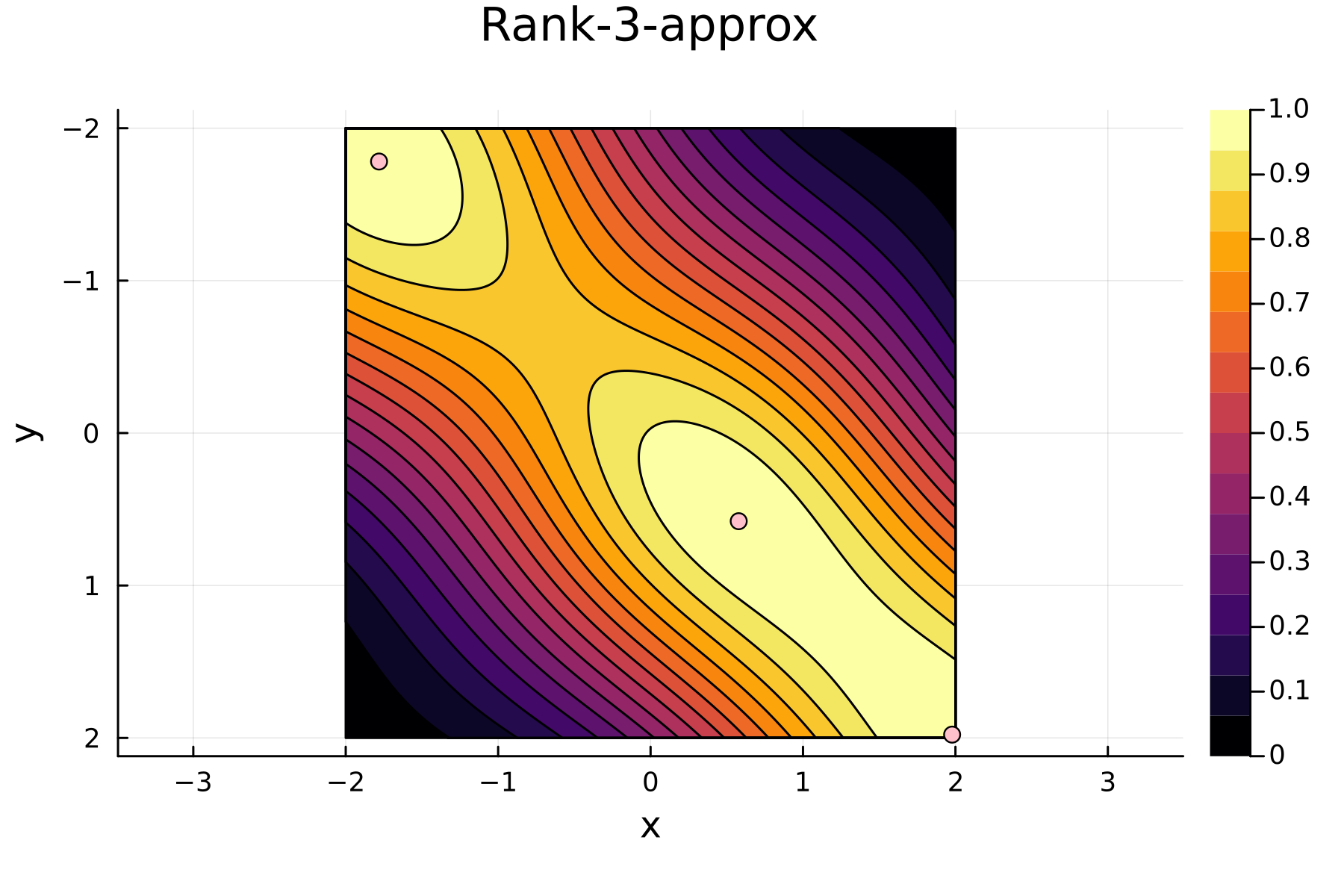
4th iteration
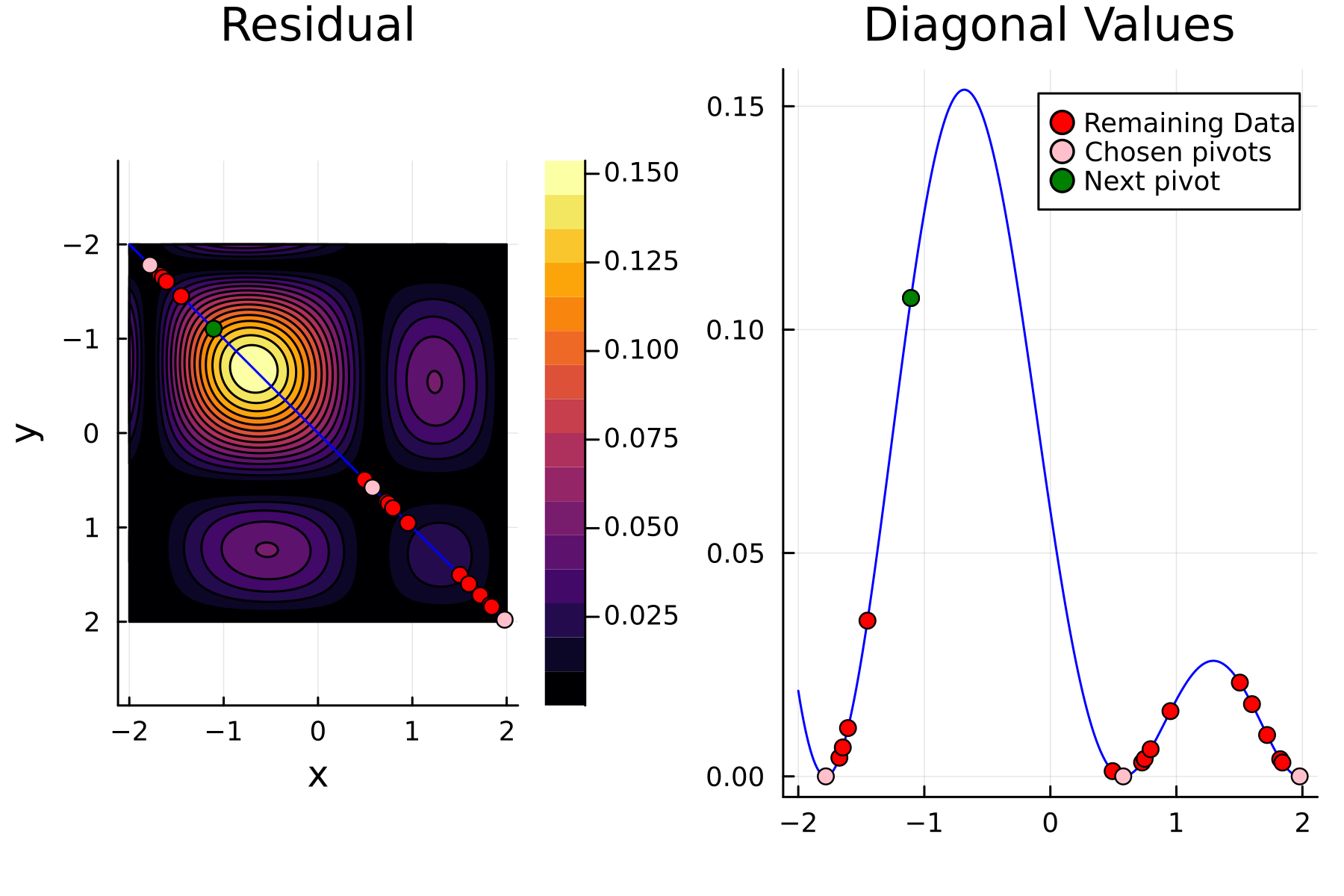
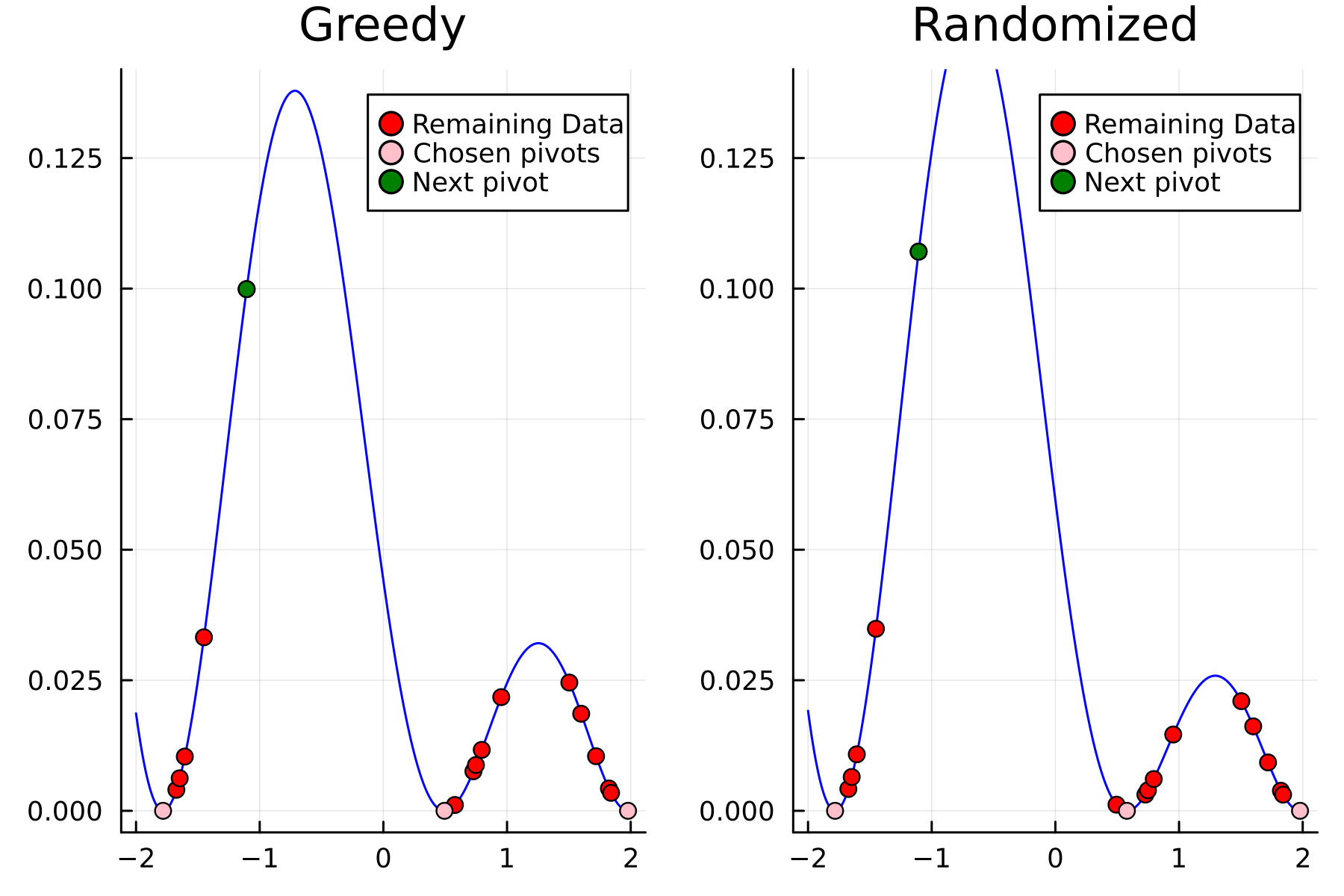
Comparing diagonals
To highlight the differences between the two approaches we plot the diagonals together. The overall decay of the diagonal is similar for each method. However, the residual within the dataset is clearly smaller for the randomized approach as compared to the greedy approach. A downside of the randomized approach is that the approximation is not as good “outside” of the data. Said differently, the randomized approach generalizes worse than the greedy approach, by relying on pivots inside, rather than the border, of the dataset.
Final remarks
A thing to note about the above is that while the approach only uses pivot points that are in dataset, this not a requirement. That is, a better pivot point could be found outside the dataset. This would be similar to the inducing point approach in sparse Gaussian processes